Gut Check: Evolution Shapes the Microbiomes of Warblers
Probiotic products like yogurt and kombucha aim to improve our well-being by cultivating the right microbes. But new research on warblers suggests that, at least in birds, "gut flora" may owe more to evolution than to ingestion.
June 28, 2023
From the Summer 2023 issue of Living Bird magazine. Subscribe now.
There has been a lot of medical news in recent years about the importance of a healthy gut microbiome for humans. Recent studies have linked the beneficial bacteria in our guts to everything from immune function to mental health, and grocery store shelves are stocked with probiotic products from yogurt to kombucha to sauerkraut that claim they’ll help us cultivate the right microbes to improve our well-being.
Birds host bacteria in their guts, too, and a new study published in the journal Molecular Ecology analyzed bacterial DNA in bird poop to look at what drives differences among the gut microbiomes of more than a dozen warbler species. The results show that evolution appears to be the biggest factor, opening up new research questions about how bird species diverge and adapt.
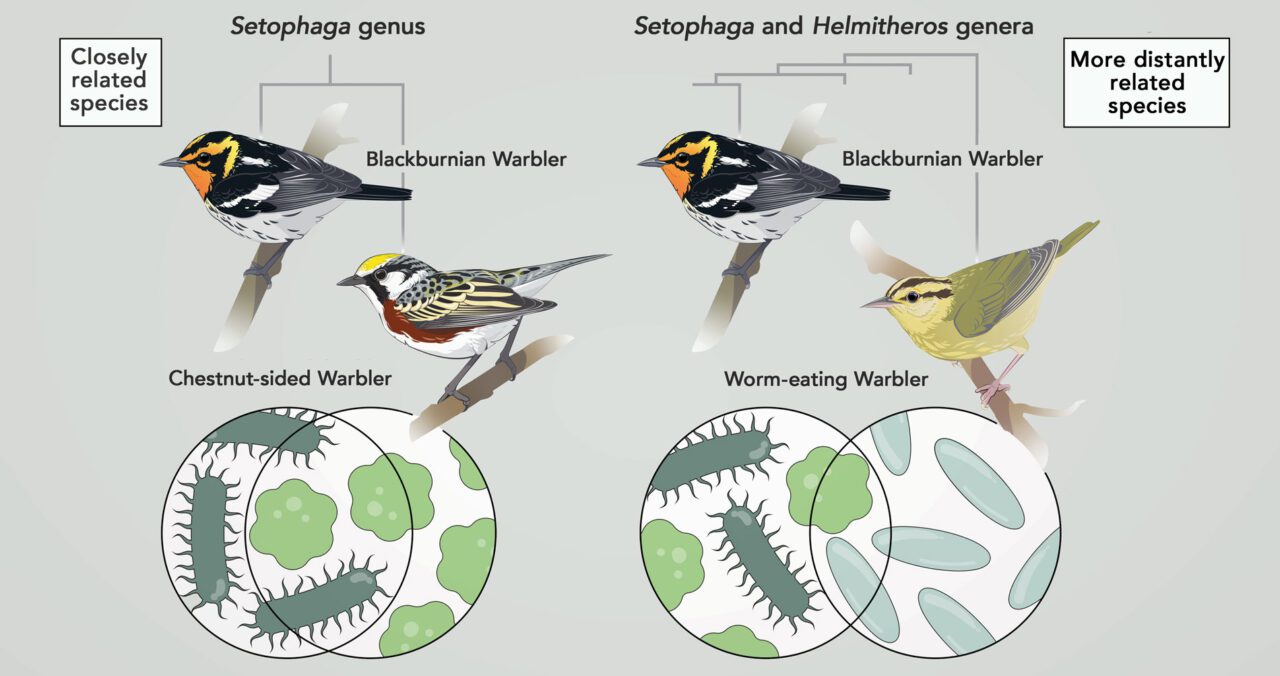
Past studies in mammals have found that the average makeup of gut-bacteria communities across species is likely to be similar among individuals from closely related species, and less similar among more distant relatives. In birds, however, this pattern has been harder to detect. Compared to mammals, birds have short, simple digestive tracts—an adaptation for flight—and food passes through their bodies quickly, potentially leading to lots of turnover in the types of bacteria they harbor.
“The conventional wisdom is that the microbiomes of birds are not all that exciting—there’s a lot of individual variation, but not a lot of predictability amongst species,” says study coauthor David Toews, a biology professor at Penn State University.
In 2017, Toews (then a Cornell Lab of Ornithology postdoctoral researcher) teamed up with Eliot Miller (a current postdoctoral associate at the Cornell Lab) to begin collecting fecal samples from 15 warbler species in New York’s Adirondacks. They intended to analyze the fragments of DNA present in the samples to study the foraging and diets of warblers. Miller describes their Adirondack fieldwork as “simultaneously gorgeous and a little bit atrocious. The bugs are horrible.”

Miller and Toews camped in the southwestern Adirondacks, driving down dirt roads in pursuit of their target species and playing recorded calls to lure territorial males into mist nets.
“We’d get up really early, make some coffee, have a quick breakfast, hop in the car, and strategize about how we were going to catch the species on our checklist for the day,” Miller says. The team visited specific microhabitats where they had the best chance to catch species such as Chestnut-sided Warblers, Black-throated Green Warblers, and Common Yellowthroats.
To collect a fecal sample, they would simply place a captured warbler inside a paper bag and wait for it to defecate before removing it and banding it.


“I was skeptical when we were first doing it,” admits Toews. “I wasn’t sure they were really going to, you know, create a fecal sample on command. But 99% of birds, within five minutes, will leave something in the bag.”
The original plan was to use the samples to study what insects the birds had been eating. However, when evolutionary genomics researcher Marcella Baiz joined Toews’s lab at Penn State as a postdoctoral researcher in 2019, the aims of the project expanded. Baiz, who had previously used fecal samples to study the genetics of howler monkeys, was interested in another category of organism with DNA traces that could be found in poop: gut microbes. Ultimately, she hopes to study how gut microbiomes shift as host species evolve and diverge.
The team expanded their fieldwork to include a site in Pennsylvania in 2019 and 2020, ending up with samples from 408 birds. Then Baiz headed to the lab to begin DNA analysis.
“The lab side of this work is very tedious. I find it less enjoyable [than the fieldwork] in many ways,” she says. “It’s really difficult to work with fecal samples. The DNA is really degraded, so there’s lots of steps in the protocol to try to get a quality DNA sample [out of feces].”
The technique Baiz used is called metabarcoding, a method that lets her prepare fecal samples for DNA analysis and, after careful cleaning and processing, identify some of the DNA sequences in the gut-bacteria species that are present. By targeting the specific sequences of DNA in different taxonomic groups, Baiz was able to identify 39 different phyla of gut bacteria. (In taxonomy, distinct phyla are groups of organisms that can be as different from each other as earthworms and mollusks and vertebrates.)



The resulting study—published with Baiz as lead author, and Miller and Toews as coauthors—found that the host species’ evolutionary relationships had the greatest effect on the makeup of each bird’s gut microbiome. In other words, the warblers of closely related species were most likely to have similar gut microbe communities.
“It’s rare to find a strong host phylogeny effect [an effect of hosts’ evolutionary relationships] on the microbiome in birds, so this is very new and exciting data,” says University of Alaska Anchorage postdoctoral fellow Kirsten Grond, an expert in avian gut microbiomes who was not involved in this study. “Using metabarcoding to identify the diet of the warblers is a great method to address this issue,” due to the difficulty of determining wild birds’ diets through observation alone.
The study findings published in Molecular Ecology lay the groundwork for the questions Baiz hopes to study next. For example, if the microbiome communities of birds diverge when one species splits into two during evolution, she wonders: “What are the implications of that for the evolutionary processes that the host is undergoing?” Are birds’ microbiomes simply drifting apart as different lineages become isolated from each other, or “are the microbial communities actually coevolving with the host?” She has since launched a follow-up project to examine the gut microbiomes of hybridizing pairs of warbler species, such as Golden-winged and Blue-winged Warblers.
Baiz says that there are many avenues for future research in avian microbiomes, because scientists have very little understanding of what role gut bacteria play in digestion and health for wild birds.
“The field of microbiome research is so new, even in humans,” says Baiz, “and it’s a really hard thing to study, especially in non-model organisms [organisms that have not already received intensive research] like warblers.”
One Mystery Gut Microbe May Power the Final Leg of Blackpoll Warbler Migration
By Gustave Axelson

Another study of warbler gut bacteria—this one focusing on a single species, the Blackpoll Warbler—documented an extensive changeover in their microbiome during fall migration.
Cornell Lab of Ornithology Rose Postdoctoral Fellow Brian Trevelline started the study based on a hypothesis that Blackpoll Warblers are morphing their bodies both inside and out during their epic intercontinental flights between breeding grounds in Canada and Alaska and overwintering areas in South America.
“These birds go through amazing physiological transformations during migration,” Trevelline says, noting that Blackpolls are able to double their body mass and shrink the size of their livers and other organs to conserve energy. “Their organs are remodeling. What’s stopping their microbiomes from remodeling, too?”
Past research published in the journal Molecular Ecology has shown that the gut bacteria of Kirtland’s Warblers on their breeding grounds in Michigan differ from their gut bacteria on wintering grounds in the Bahamas. Other research recently published in the same journal showed that the microbiomes of four species of thrushes were changing during migration.
For this study, which is currently in peer review, Trevelline partnered with nine research sites across Canada and the United States to get fecal samples from Blackpoll Warblers that were collected while the birds were in a paper bag, awaiting banding. For one aspect of the study, Trevelline focused on samples collected from blackpolls in a region from the Maritime Provinces to the Carolinas—their halfway-point pit stop during fall migration, where they spend about two weeks fattening up for the final grueling leg of their journey, a nonstop flight over several days and 2,000 miles of open ocean to their South American wintering grounds.
Trevelline and colleagues on the research sequenced all of the DNA present in the fecal samples. They found that, when compared to samples collected from breeding-season blackpolls in western Canada, the gut bacteria of fall-migrating blackpolls had more genes related to functions that could benefit birds during energy-intensive feats of long-distance flight, such as genes for synthesizing amino acids (for muscle building) and metabolizing carbohydrates (for energy).
What’s more, Trevelline found that the composition of gut bacteria present in these migrating blackpolls was almost completely different than fecal samples from breeding blackpolls. In fact, the microbiomes of blackpolls on fall migration primarily consisted of just one species of bacteria.
And, Trevelline says, that microbe species is unknown to science—heightening the mystery.
But it turns out undescribed gut bacteria aren’t unusual in these kinds of studies.
“A large portion of the avian microbiome has never been described,” he says. That’s why Trevelline is eager to keep exploring the gut bacteria of birds. “It’s a new frontier of avian microbiology.”

All About Birds
is a free resource
Available for everyone,
funded by donors like you
American Kestrel by Blair Dudeck / Macaulay Library